Sources: https://github.com/deepmind/alphafold
License: https://github.com/deepmind/alphafold/blob/main/LICENSE (non-commercial use only)
Note: Any publication that discloses findings arising from using this source code or the model parameters should cite the AlphaFold paper.
Genetic Databases: /beegfs/desy/group/it/ReferenceData/alphafold
Samples: /software/alphafold/2.0
Docker image: gitlab.desy.de:5555/frank.schluenzen/alphafold
Singularity (docker) image: docker://gitlab.desy.de:5555/frank.schluenzen/alphafold:sif
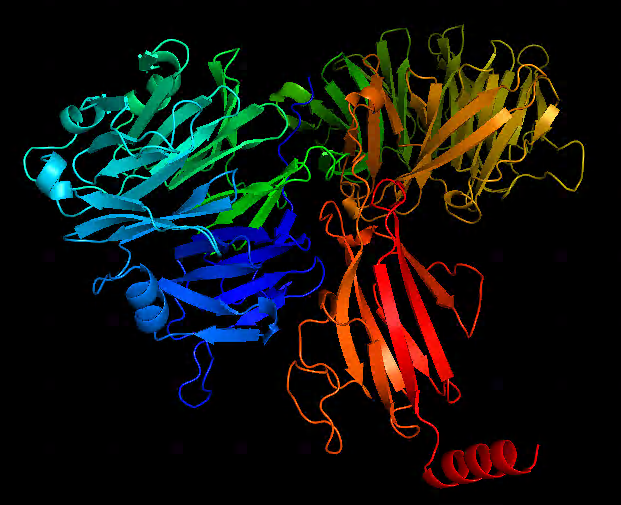
casp14: https://www.predictioncenter.org/casp14/index.cgi (T1050)
This page briefly describes the installation of AlphaFold v2.0 on Maxwell.
AlphaFold needs multiple genetic (sequence) databases to run. The databases are huge (2.2TB). We therefore provide a central installation under /beegfs/desy/group/it/ReferenceData/alphafold.
Samples and sources for alphafold can be found in /software/alphafold/2.0.
Below are descriptions how to run alphafold using docker resp. singularity. Both installations have been tested to some extend, and don't differ much in usability, so feel free to choose, or to use your own images or local installation.
Content
Running alphafold using docker
The alphafold documentation proposes to use dockers python interface to run the alphafold image. This turns out to be a bit cumbersome in our environment, and in the end seems only to add a few variables to the runtime environment. Alternatively there are simple bash-scripts provided to run alphafold interactively or as a batch job.
You should be able to simply run the script with the following command:
AF_outdir=/beegfs/desy/user/$USER/ALPHAFOLD /software/alphafold/2.0/alphafold.sh --fasta_paths=/software/alphafold/2.0/T1050.fasta --max_template_date=2020-05-1
- The default output-directory is /tmp/alphafold. In batch-jobs this folder is removed at the end of the job, so you have to make sure to copy it somewhere beforehand, or simply define AF_outdir as shown.
- You would of course need to alter the fasta-sequence and possibly the template-date.
- You can override almost all variables used in the script above.
There is also a template batch-script which you could copy and adjust according to your needs:
#!/bin/bash #SBATCH --partition=maxgpu #SBATCH --constraint='A100|V100' #SBATCH --time=0-12:00 export AF_outdir=/beegfs/desy/user/$USER/ALPHAFOLD mkdir -p $AF_outdir /software/alphafold/2.0/alphafold.sh --fasta_paths=/software/alphafold/2.0/T1050.fasta --max_template_date=2020-05-1
|
Running alphafold using singularity
The sample script for running alphafold inside a singularity container is almost identical to the docker-version.
You should be able to simple run the script with the following command:
AF_outdir=/beegfs/desy/user/$USER/ALPHAFOLD /software/alphafold/2.0/alphafold-singularity.sh --fasta_paths=/software/alphafold/2.0/T1050.fasta --max_template_date=2020-05-1
- The default output-directory is /tmp/alphafold. In batch-jobs this folder is removed at the end of the job, so you have to make sure to copy it somewhere beforehand, or simply define AF_outdir as shown.
- You would of course need to alter the fasta-sequence and possibly the template-date.
- You can override almost all variables used in the script above.
There is also a template batch-script which you could copy and adjust according to your needs:
#!/bin/bash #SBATCH --partition=maxgpu #SBATCH --constraint='A100|V100' #SBATCH --time=0-12:00 export SINGULARITY_CACHEDIR=/beegfs/desy/user/$USER/scache export TMPDIR=/beegfs/desy/user/$USER/stmp mkdir -p $TMPDIR $SINGULARITY_CACHEDIR export AF_outdir=/beegfs/desy/user/$USER/ALPHAFOLDS /software/alphafold/2.0/alphafold-singularity.sh \ --fasta_paths=/software/alphafold/2.0/T1050.fasta --max_template_date=2020-05-1
|
Building the docker image
You essentially just need to follow the instructions at https://github.com/deepmind/alphafold. The image used in the sample was built roughly like this:
mkdir -p /beegfs/desy/user/$USER/scratch cd /beegfs/desy/user/$USER/scratch git clone https://github.com/deepmind/alphafold.git cd alphafold cp /software/alphafold/2.0/docker/run_docker.py docker/ # or modify the script to point to the db-directory docker login gitlab.desy.de:5555 docker build -t gitlab.desy.de:5555/frank.schluenzen/alphafold -f docker/Dockerfile . docker push gitlab.desy.de:5555/frank.schluenzen/alphafold
For singularity we needed a small modification (see also https://github.com/deepmind/alphafold/issues/32):
mkdir -p /beegfs/desy/user/$USER/scratch cd /beegfs/desy/user/$USER/scratch git clone https://github.com/deepmind/alphafold.git cd alphafold cp /software/alphafold/2.0/docker/run_docker.py docker/ # or modify the script to point to the db-directory # there is a copy of stereo_chemical_props.txt in /beegfs/desy/group/it/ReferenceData/alphafold/ # the folder has to present in the container anyway to make use of pre-installed dbs. perl -pi -e 's|alphafold/common/stereo_chemical_props.txt|/beegfs/desy/group/it/ReferenceData/alphafold/stereo_chemical_props.txt|' alphafold/common/residue_constants.py docker login gitlab.desy.de:5555 docker build -t gitlab.desy.de:5555/frank.schluenzen/alphafold:sif -f docker/Dockerfile . docker push gitlab.desy.de:5555/frank.schluenzen/alphafold:sif
Runtime
| CPU | Cores | Memory | GPU | #GPU | Image | Elapsed | |
|---|---|---|---|---|---|---|---|
| 1 | AMD EPYC 7302 | 2x16 (+HT) | 512GB | NVIDIA A100-PCIE-40GB | 4 | singularity | 3:49:07 |
| AMD EPYC 7302 | 2x16 (+HT) | 512GB | NVIDIA A100-PCIE-40GB | 4 | docker | 4:04:03 | |
| 2 | Intel(R) Xeon(R) Silver 4216 | 2x16 (+HT) | 384GB | NVIDIA Tesla V100-PCIE-32GB | 1 | docker | 5:28:55 |
| Intel(R) Xeon(R) Silver 4216 | 2x16 (+HT) | 384GB | NVIDIA Tesla V100-PCIE-32GB | 1 | singularity | 5:02:50 | |
| 3 | Intel(R) Xeon(R) Gold 6240 | 2x18(+HT) | 384GB | NVIDIA Tesla V100-SXM2-32GB | 4 | singularity | 4:46:45 |
Intel(R) Xeon(R) Gold 6240 | 2x18(+HT) | 384GB | NVIDIA Tesla V100-SXM2-32GB | 4 | docker | 5:06:43 | |
| 4 | Intel(R) Xeon(R) E5-2640 v4 | 2x10(+HT) | 256GB | Tesla P100-PCIE-16GB | 1 | singularity | 7:23:26 |
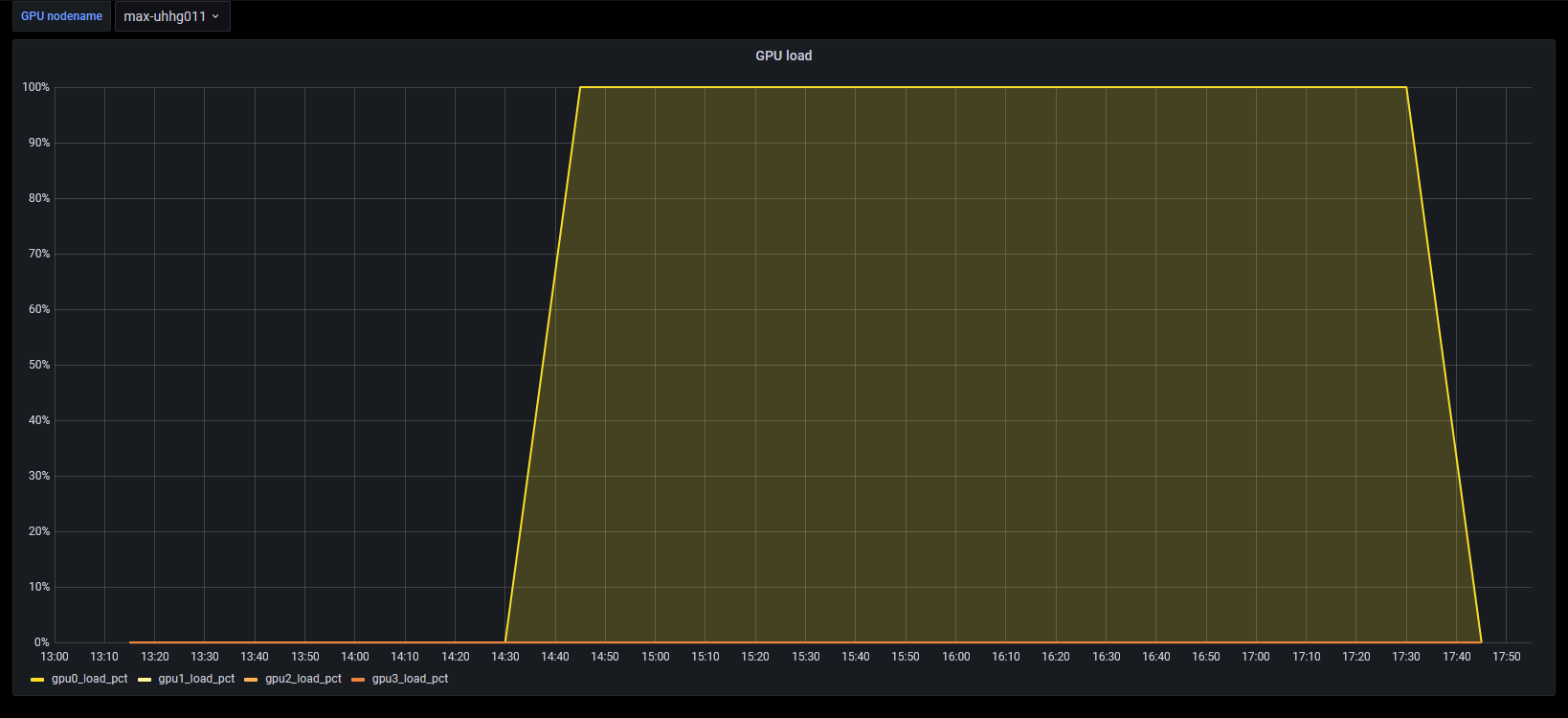
Utilization
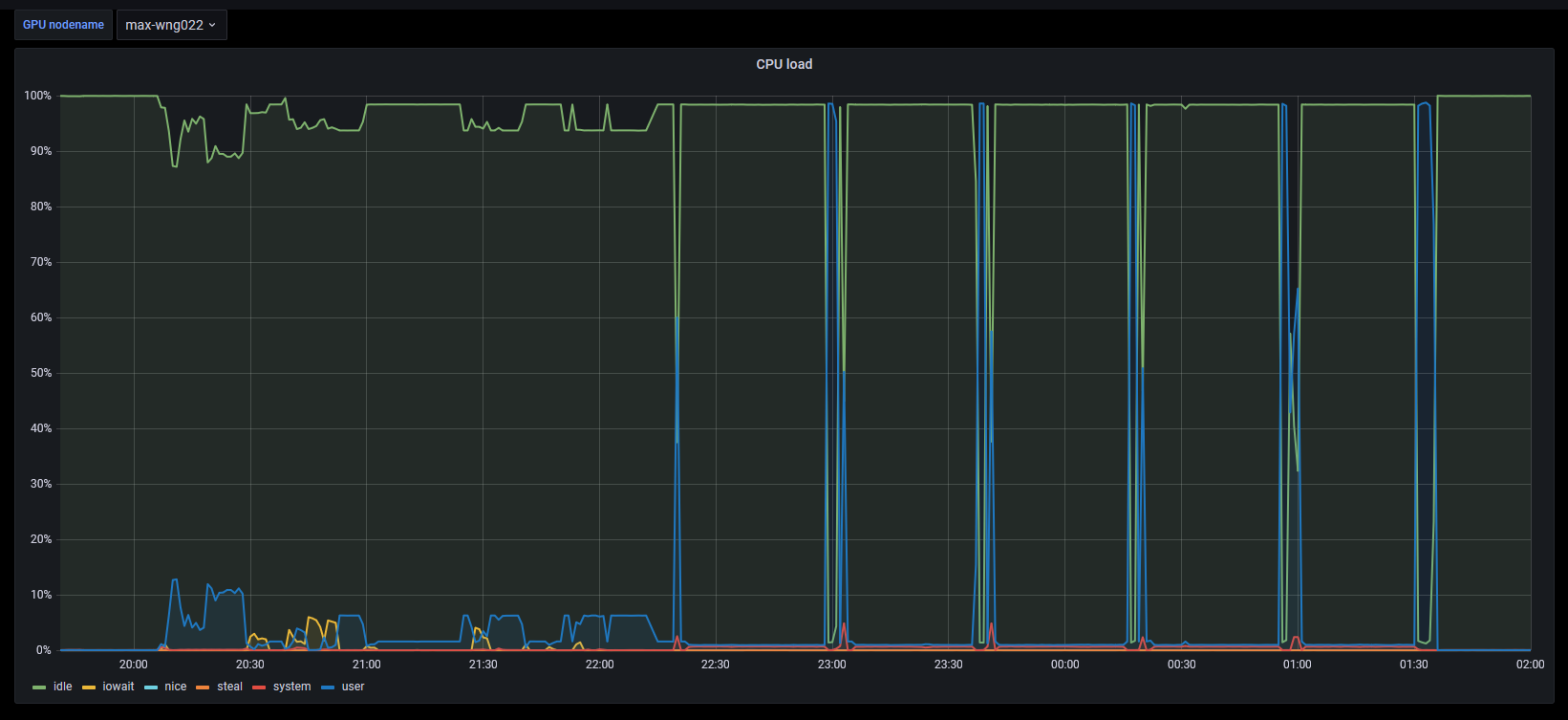
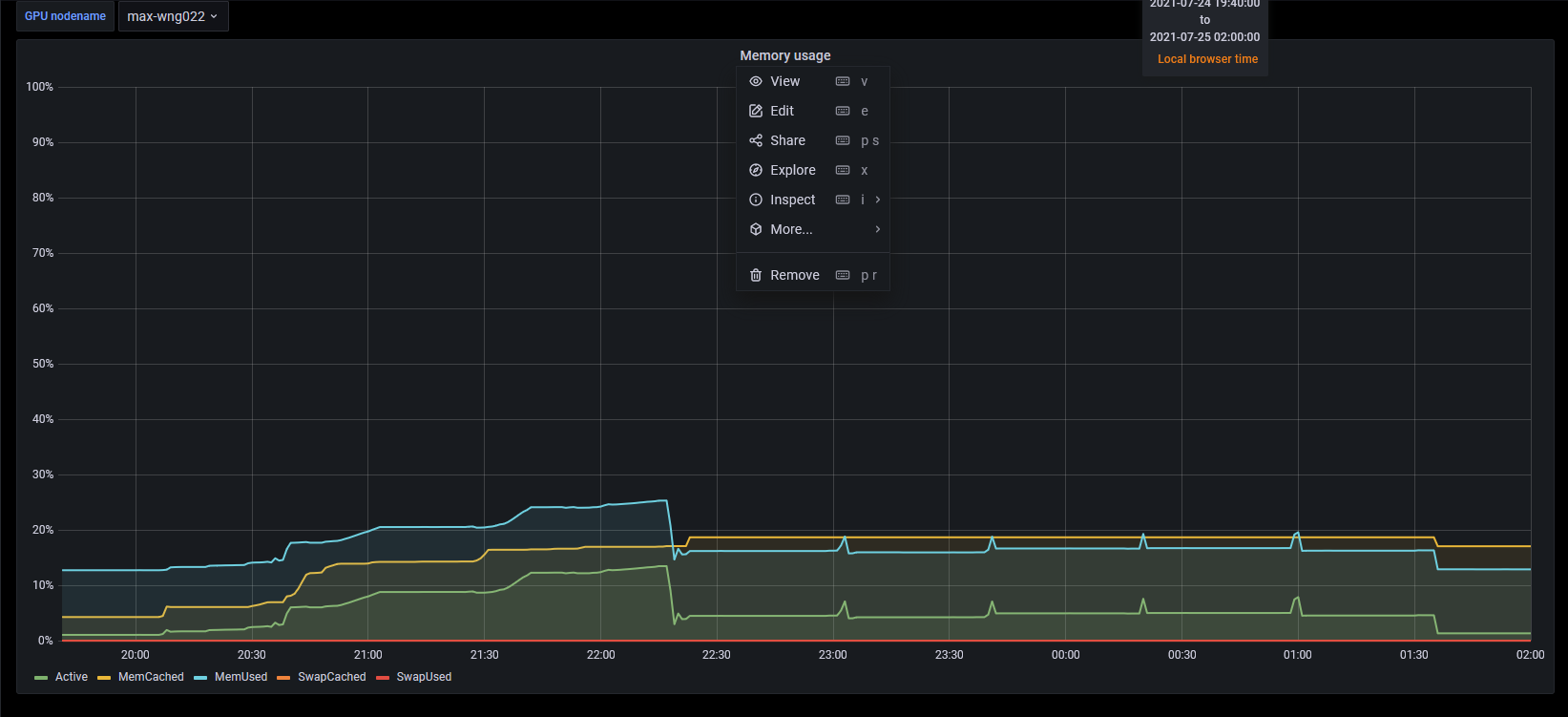
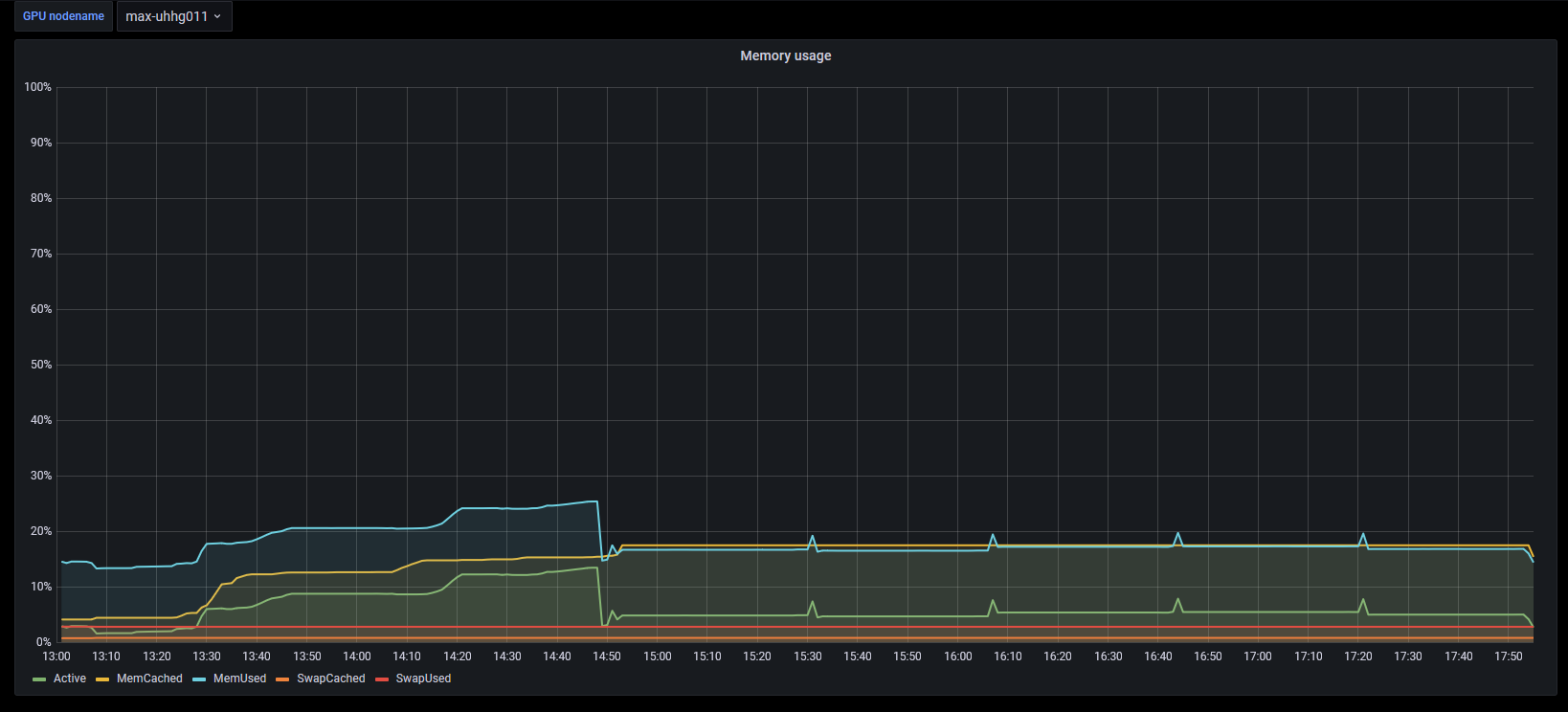
The use pattern of CPU and GPU looks a little peculiar.
| # | CPU | Memory | GPU | GPU memory |
|---|---|---|---|---|
| 2 | ||||
| 3 |
Remarks
- An alphafold clone can be found under https://gitlab.desy.de/frank.schluenzen/alphafold. CI/CD has been set up. The pipeline jobs claim to be successful, but actually die silently half way through the build. So it might currently not be possible to build the docker images automatically.
- Both docker and singularity have successfully been tested with an unprivileged account and without usernamespaces. It's however quite possible that the installations are very far from being perfect. Drop a message to maxwell.service@desy.de if you observe any problems (or have a better setup).
- Running the images you'll see messages like Unable to initialize backend 'tpu': Invalid argument: TpuPlatform is not available. These messages just warn that Nvidia A100/V100 is not a TPU (e.g. https://github.com/deepmind/alphafold/issues/25).
- There is no indication that alphafold uses more than 1 GPU for computations (see also https://github.com/deepmind/alphafold/issues/30 though it's not very explicit). It should however be able to use unified memory, so being able to utilize memory of additional GPUs as well as CPU memory. The threads indicates that setting TF_FORCE_UNIFIED_MEMORY=1, XLA_PYTHON_CLIENT_MEM_FRACTION=4.0 might be necessary to achieve that, but it's certainly not benefitial for models fitting into memory of a single GPU (and probably only makes sense on nvlink nodes).
- Singularity has certain advantages (e.g. operates entirely in user space and has good nvidia support) and RedHat meanwhile dropped Docker support for RedHat/Centos 8. However, alphafold is going for docker, and none-docker installations tend to run into problems, so might give a smoother experience. Surprisingly the singularity runs appear to be significantly faster than docker.