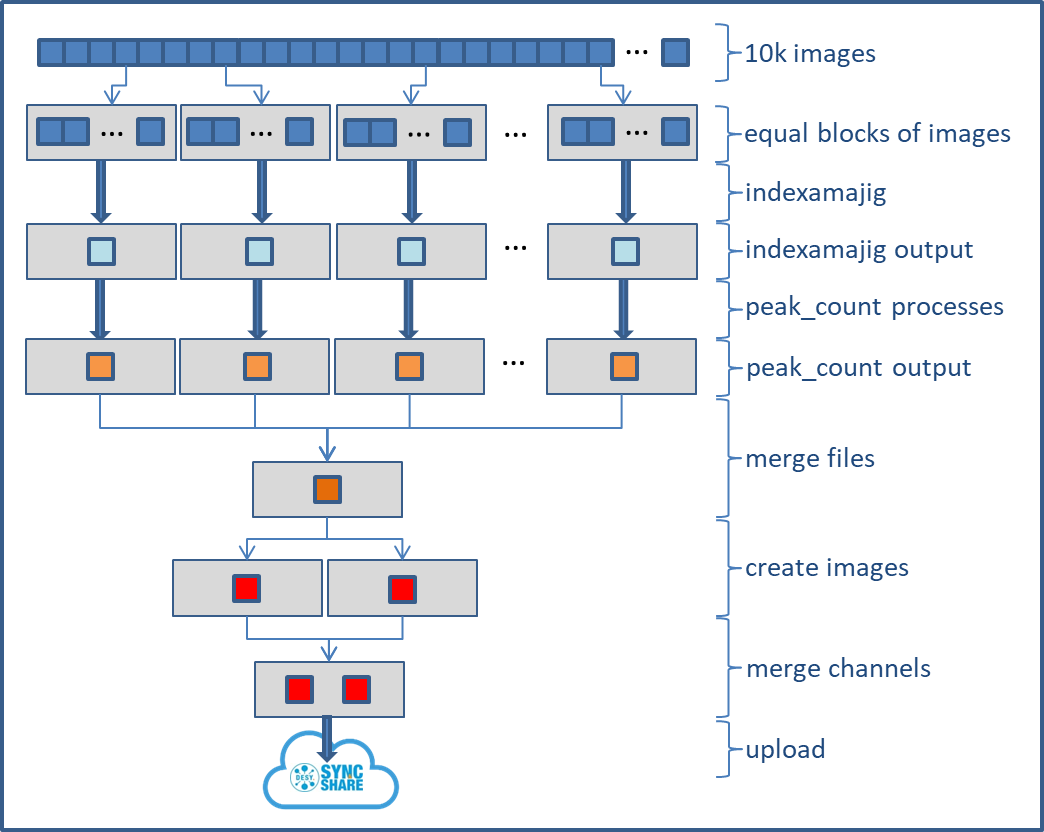
The job submission script
A sample submission script could look like this:
In detail
The first part of the submission script just sets the environment. Please note that the "base job" allocates only a single node!
environment
#!/bin/bash #SBATCH --time=0-01:00:00 # you need just a single node, kind of a master which orchestrates the jobs #SBATCH --nodes=1 #SBATCH --partition=upex #SBATCH --job-name=swift-split #SBATCH --dependency=singleton # I've chosen a name which reflects the number of nodes (58) to use #SBATCH --output=swift-split-58.out #SBATCH --tasks-per-node=1 # that's just to ensure that all jobs run on identical nodes for benchmarking #SBATCH --constraint=Gold-6240 # the basic setup export LD_PRELOAD="" source /etc/profile.d/modules.sh module load maxwell swift
The rest is the preparation of the input file, and running the actual workflow:
preparation & environment
# it's easier to embed individual tasks into small scripts, and it's easier to have those in the PATH basedir=/beegfs/desy/user/schluenz/Crystfel.Swift export PATH=$basedir:$PATH # pass constraints and limits to slurm. It's also possible to declare those in the site config: export SBATCH_CONSTRAINT=Gold-6240 export SBATCH_TIMELIMIT=02:00:00 # use 58 nodes, so split the list of images into 58 roughly equal parts numnodes=58 mkdir -p $basedir/procdir # collect a list of images and count them ls -1 /beegfs/desy/group/it/ReferenceData/cxidb/ID-21/cxidb-21-run01[34]*/data1/LCLS*.h5 > $basedir/procdir/files.to.process num_images=$(ls -1 /beegfs/desy/group/it/ReferenceData/cxidb/ID-21/cxidb-21-run01[34]*/data1/LCLS*.h5 | wc -l) # split the files splitlines=$(( $(cat $basedir/procdir/files.to.process | wc -l) / $numnodes )) /usr/bin/split -l $splitlines -d $basedir/procdir/files.to.process $basedir/procdir/xxx # run the workflow swift -sites upex -config maxwell.conf indexamajig.swift -nsplit=$numnodes
the workflow
type file;
# this generates just a list of images for each of the 58 parts and puts them into new set of files. There are simpler ways of doing that...
# splitter is the name of the scriplet. Note: you can't have '.' in your filename!
app (file o) split (int i, int nsplit)
{
splitter i nsplit stdout=filename(o);
}
# actually run indexamajig.
app (file o) indexamajig (int i)
{
indexamajigwrap i stdout=filename(o);
}
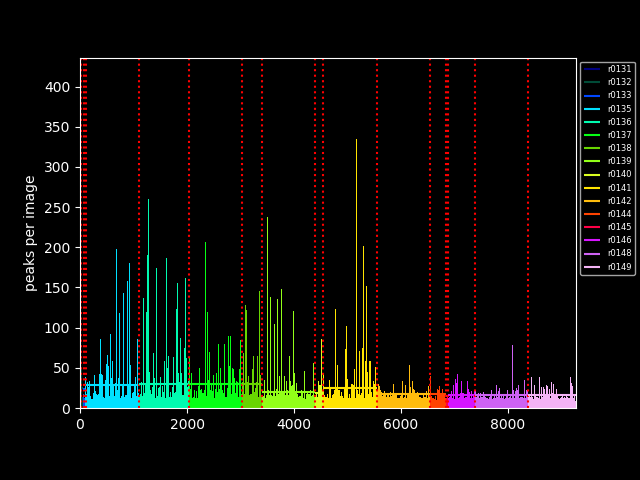
# collect information about number of peaks per image
app (file o) peak_count (file infile)
{
peak_count filename(infile) stdout=filename(o);
}
# ... and create a plot for peak counts
app (file o, file image) plot_peaks (file infile)
{
plot_peaks filename(infile) filename(image) stdout=filename(o);
}
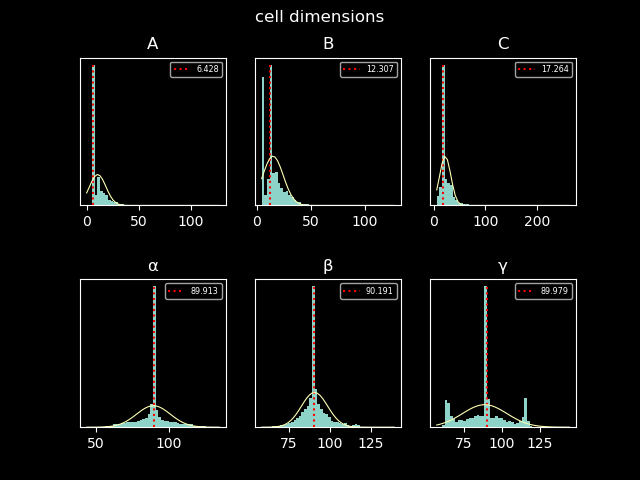
# ... and cell dimension
app (file o, file image) plot_cell (file infile)
{
plot_cell filename(infile) filename(image) stdout=filename(o);
}
# merge all peak information into a single file
app (file o) merge_peaks (file s[])
{
cat filenames(s) stdout=filename(o);
}
# if nsplit is not declared on the command line default to 16 nodes
int nsplit = toInt(arg("nsplit","16"));
file image_list[];
file peaks[];
foreach i in [0:nsplit-1] {
file splitout <single_file_mapper; file=strcat("output/split_",i,".out")>;
file indexamajigout <single_file_mapper; file=strcat("output/indexamajig_",i,".out")>;
file peak_countout <single_file_mapper; file=strcat("output/peak_count_",i,".out")>;
# create the lists of images
splitout = split(i, nsplit);
image_list[i] = splitout;
# process all sets of images. Though it looks serial it's actually done in parallel
indexamajigout = indexamajig(i);
# get number of peaks from indexamajib processing. With indexamajigout being the output of indexamajig and input for peak_counting, swift will schedule the job only when indexamajig is done.
peak_countout = peak_count(indexamajigout);
peaks[i] = peak_countout;
}
# combine the peak counting stats into a single file
file all_peaks <single_file_mapper; file=strcat("output/peaks.out")>;
all_peaks = merge_peaks(peaks);
# create two plots. again that's actually processed in parallel.
file cimage <"/beegfs/desy/user/schluenz/Crystfel.Swift/output/cells.png">;
file pimage <"/beegfs/desy/user/schluenz/Crystfel.Swift/output/peaks.png">;
file pp <single_file_mapper; file=strcat("output/pp.out")>;
file pc <single_file_mapper; file=strcat("output/pc.out")>;
(pc,cimage)=plot_cell(all_peaks);
(pp,pimage)=plot_peaks(all_peaks);