The job submission script
A sample submission script could look like this:
In detail
The first part of the submission script just sets the environment. Please note that the "base job" allocates only a single node!
environment
#!/bin/bash #SBATCH --time=0-02:00:00 # only one node is needed. The base job will then submit $numnodes child jobs, see below: #SBATCH --nodes=1 #SBATCH --partition=all #SBATCH --job-name=nextflow # just to avoid that benchmark jobs interfere with each other: #SBATCH --dependency=singleton #SBATCH --output=nextflow.out # this constraint is not really needed: #SBATCH --constraint=Gold-6240 # when submitting from max-display suppress the LD_PRELOAD warnings: export LD_PRELOAD="" # I want to use DSL 2.0 so use a newer version: export NXF_VER=19.09.0-edge export NXF_CLUSTER_SEED=$(shuf -i 0-16777216 -n 1) export NXF_ANSI_LOG=false export NXF_ANSI_SUMMARY=false # for benchmarks: make sure to run all jobs on identical hardware export SBATCH_CONSTRAINT=Gold-6240 # environment for the processes to run source /etc/profile.d/modules.sh module load mpi/openmpi-x86_64 module load crystfel
The rest is the preparation of the input file, and running the actual workflow:
preparation & workflow
# Declare the number of child jobs to be run:
numnodes=16
# define a unique name for the report written by nextflow:
report="${SLURM_JOB_NAME}-nodes-${numnodes}.report.html"
# just count how many image to process in total:
num_images=$(ls -1 /beegfs/desy/group/it/ReferenceData/cxidb/ID-21/cxidb-21-run01[34]*/data1/LCLS*.h5 | wc -l)
# split image files into $numnodes sets of equal size:
ls -1 /beegfs/desy/group/it/ReferenceData/cxidb/ID-21/cxidb-21-run01[34]*/data1/LCLS*.h5 > procdir/files.to.process
splitlines=$(( $(cat procdir/files.to.process | wc -l) / $numnodes ))
split -l $splitlines -d procdir/files.to.process procdir/xxx
# Run workflow
# - the actual workflow is defined in indexamajig.nf.split. That will be explained below
# - --in '<path-to>/procdir/xxx*' defines the input files to be processed by the workflow
# - -with-report $report we want an html report at the end of job
/software/workflows/nextflow/bin/nextflow run indexamajig.nf.split \
--in '<path-to>/procdir/xxx*' \
-with-report $report
Just for fun save the output files to desycloud:
prepare cloud upload
#
# prepare for uploads to desycloud
# - this will create a unique new folder on desycloud. You need a ~/.netrc file to upload files without username/password
#
URL="https://desycloud.desy.de/remote.php/webdav/Maxwell/nextflow/${SLURM_JOB_NAME}-nodes-${numnodes}-${SLURM_JOB_ID}"
# make sure top level folder exists:
test=$(curl -s -n -X PROPFIND -H "Depth: 1" "https://desycloud.desy.de/remote.php/webdav/Maxwell/nextflow" | grep "200 OK" | wc -l)
if [[ $test -eq 0 ]]; then
curl -s -n -X MKCOL "https://desycloud.desy.de/remote.php/webdav/Maxwell/nextflow" > /dev/null
fi
# create the new folder for future uploads
curl -s -n -X MKCOL "$URL" > /dev/null
# we will also upload some files as part of the workflow, so make sure the workflow knows the target. There are certainly better ways to do it!
perl -pi -e "s|URL=.*|URL=\"$URL\"|" indexamajig.nf.split
After all images have been processed we actually upload the nextflow report and files to run the workflow
cloud upload
#
# Upload results to desyloud
#
for file in indexamajig.nf.split nextflow-indexamajig.sh $report ; do
target=$(echo $file | rev | cut -d/ -f1 | rev)
curl -s -n -T $file "$URL/$target" > /dev/null
done
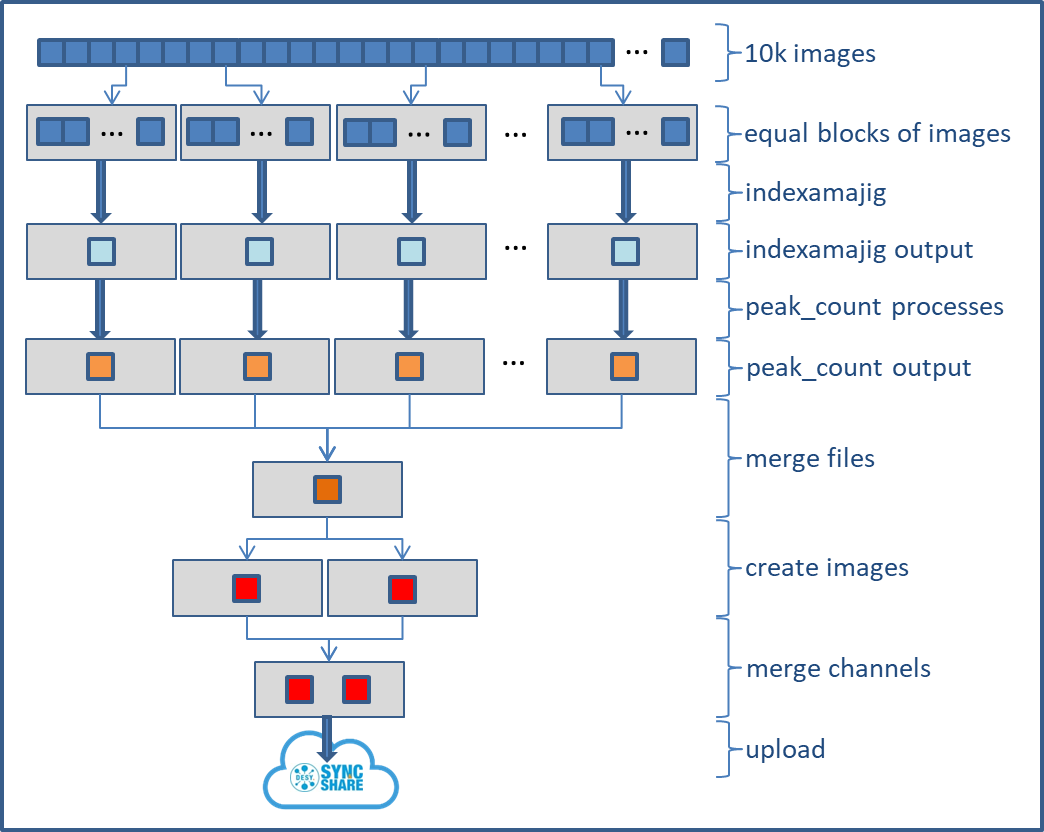
The workflow declaration
Again that's just one possible way to define the workflow to illustrate some basics. It uses version2 of the workflow description language, which I personally like much better.
indexamamjig.nf.split: the workflow declaration
/* Let's start at the end */
workflow {
/* the filenames from the script create a so called channel */
data = Channel.fromPath(params.in)
/* indexamajig function will process each of the files. The result for each of the processes will be piped into a function called peak_count */
indexamajig(data) | peak_count
/* All results are in separate files. Simply collect it in arbitrary order */
collect_index_output(indexamajig.out.collect())
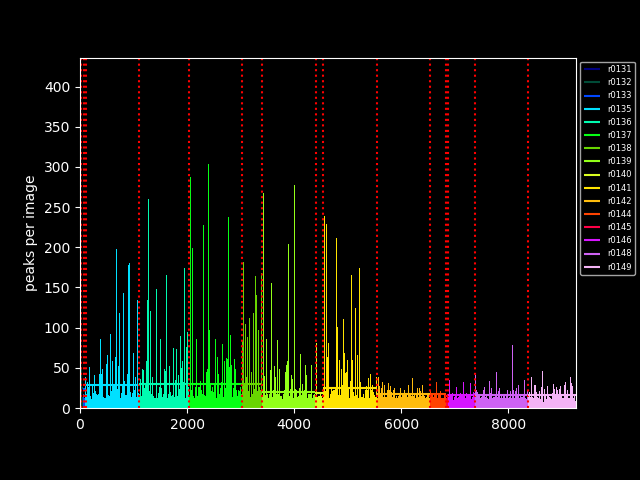
/* extract number of peaks and lattices and write to slurm log and ./outdir/peaks.info */
/* peakfile is again a channel */
peakfile=Channel.fromPath('/beegfs/desy/user/schluenz/Crystfel.Nextflow/outdir/peaks.info')
/* collect_peak info aggregates the output from peak_count runs, and pipes it into two plot routines.
the plot routines generate two images which populate two output channels. mix() merges the channels,
so that the function "upload_image" can upload all images in the channel to desycloud
*/
collect_peak_info(peak_count.out.collect()) | (simple_peak_plot & simple_cell_plot) | mix() | upload_image
}